Ensemble studies have contributed tremendously to comprehending biological reactions. However, these studies characterize the average molecular population, and have limited ability to detect intermediate states or to distinguish heterogeneities. Over the past few decades, single-molecule techniques, including fluorescence resonance energy transfer (FRET), magnetic tweezers (MT) and optical tweezers (OT), have proved to be exceedingly powerful in addressing this knowledge gap. By studying one molecule at a time, these approaches have enabled significant advancement in understanding a wide variety of biomolecular systems, especially those involving nucleic acids and associated proteins. Using these single‐molecule techniques, our research has focused on understanding mechanisms and functions of molecular motors, such as helicase, polymerase and nuclease, in the processes of DNA replication, repair, recombination, editing, and segregation and on developing technological innovations to meet the challenges in the pursuits. We are also interesed in membraneless organelles and amyloid fibrils related to neurodegenerative diseases. Currently, our lab covers five research fields:
-
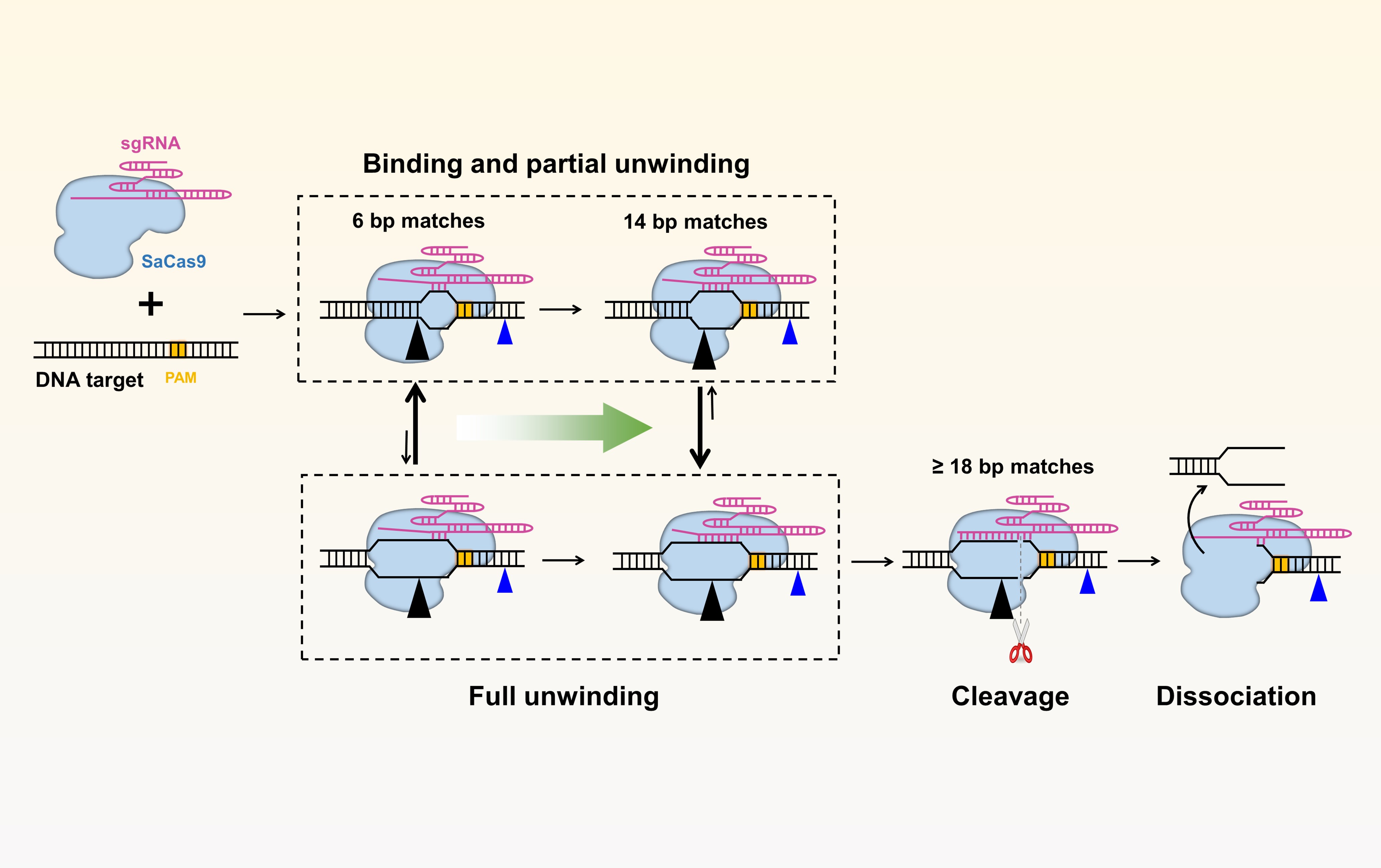 Molecular mechanisms of CRISPR-Cas proteins We use a series of single-molecule techniques to understand the detailed molecular mechanisms of CRISPR-Cas proteins, such as spCas9 and saCas9, in order to increase their fidelity, minimize off-target cleavage, and ensure their efficient applications in medicine and biology.
Molecular mechanisms of CRISPR-Cas proteins We use a series of single-molecule techniques to understand the detailed molecular mechanisms of CRISPR-Cas proteins, such as spCas9 and saCas9, in order to increase their fidelity, minimize off-target cleavage, and ensure their efficient applications in medicine and biology. -
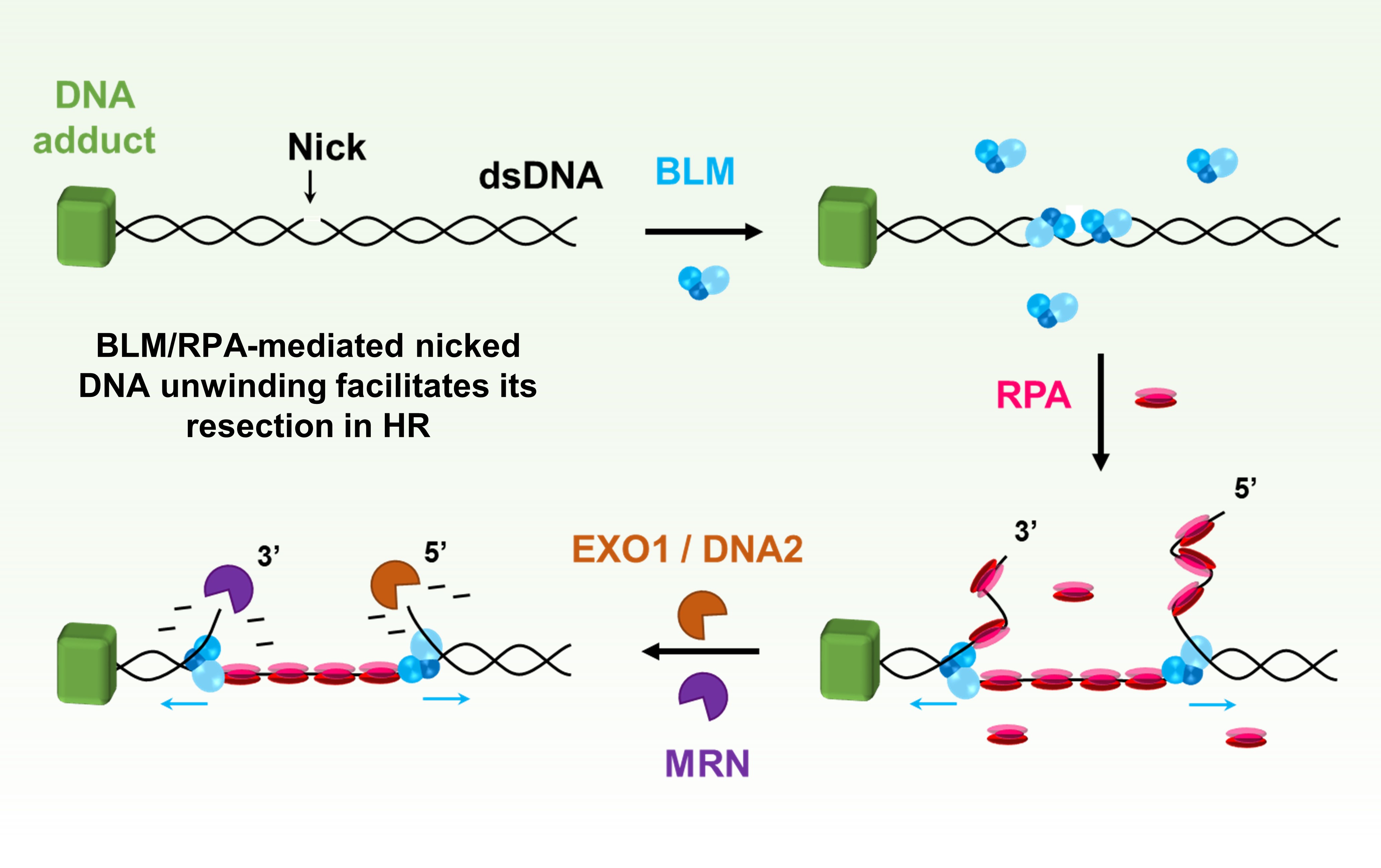 Homologous recombination-directed DSB repair DNA repair and recombination are essential for maintaining genome integrity. We combine biophysical manipulation techniques with fluorescence imaging to control, visualize, and dissect key steps in the recombination reaction mediated by many molecular proteins down to the single molecule.
Homologous recombination-directed DSB repair DNA repair and recombination are essential for maintaining genome integrity. We combine biophysical manipulation techniques with fluorescence imaging to control, visualize, and dissect key steps in the recombination reaction mediated by many molecular proteins down to the single molecule. -
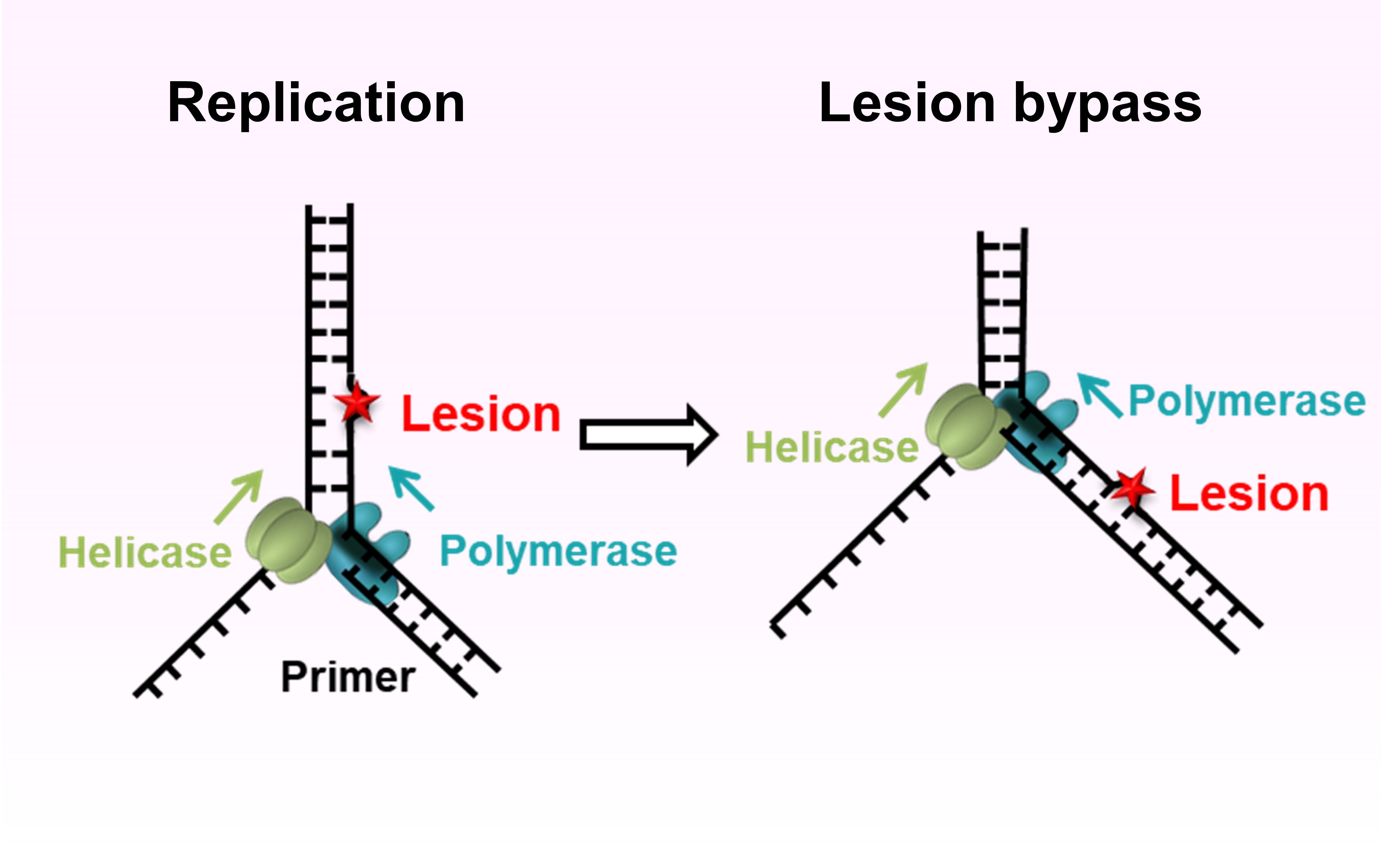 Replication restart pathways after collision with obstacles To ensure accurate DNA replication, a replisome must effectively overcome numerous obstacles on its DNA substrate. We empoly single-molecule techniques to reveal pathways that cells have evolved to possess in order to avoid replication failure from fork barriers, such as DNA damage and DNA -bound complexes.
Replication restart pathways after collision with obstacles To ensure accurate DNA replication, a replisome must effectively overcome numerous obstacles on its DNA substrate. We empoly single-molecule techniques to reveal pathways that cells have evolved to possess in order to avoid replication failure from fork barriers, such as DNA damage and DNA -bound complexes. -
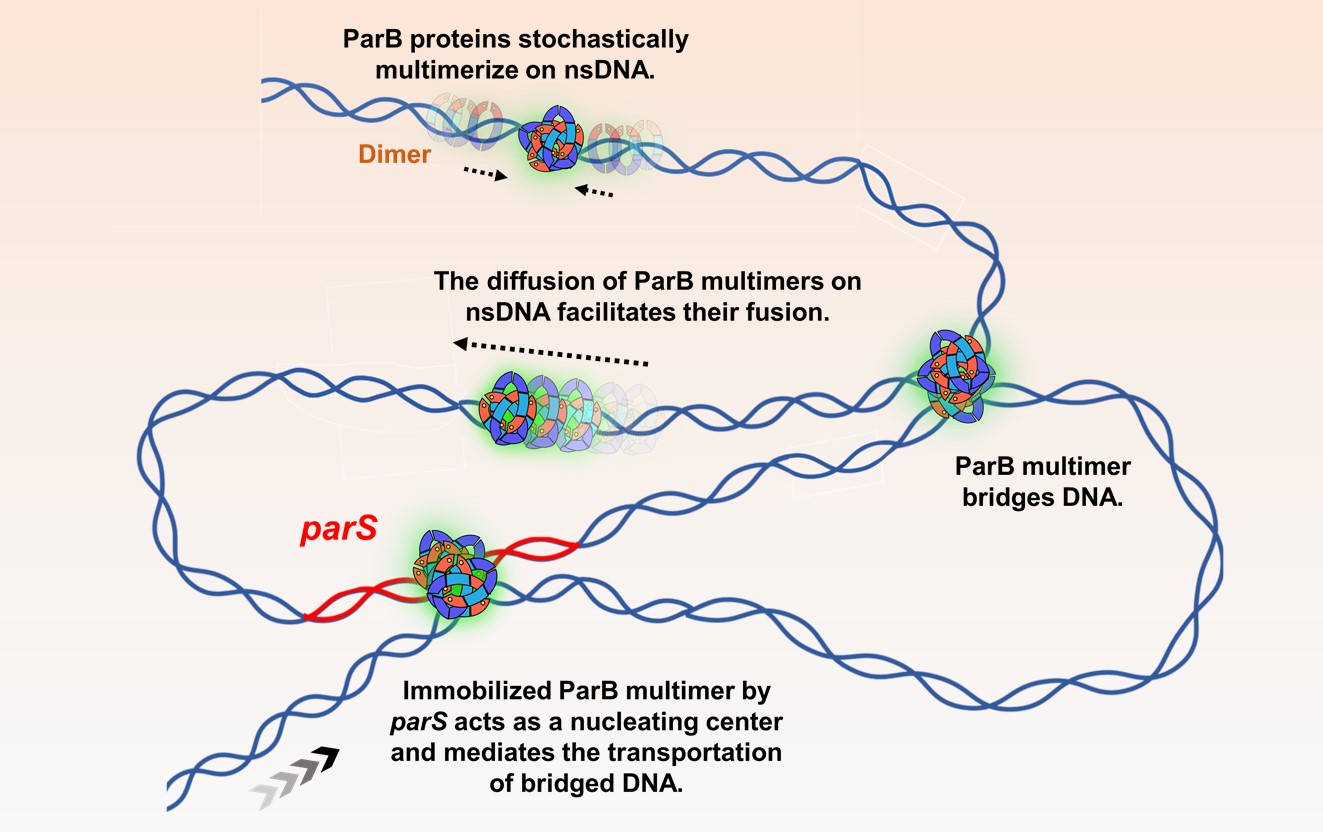 ParABS-mediated bacterial chromosome segregation The tripartite ParABS system mediates chromosome segregation in a wide range of bacteria, which ensures the inheritance of a full copy of the genetic information of the parent to each daughter cell. Using a series of single-molecule techniques, we aim to understand the organization and function of the partition complex in chromosome segregation.
ParABS-mediated bacterial chromosome segregation The tripartite ParABS system mediates chromosome segregation in a wide range of bacteria, which ensures the inheritance of a full copy of the genetic information of the parent to each daughter cell. Using a series of single-molecule techniques, we aim to understand the organization and function of the partition complex in chromosome segregation. -
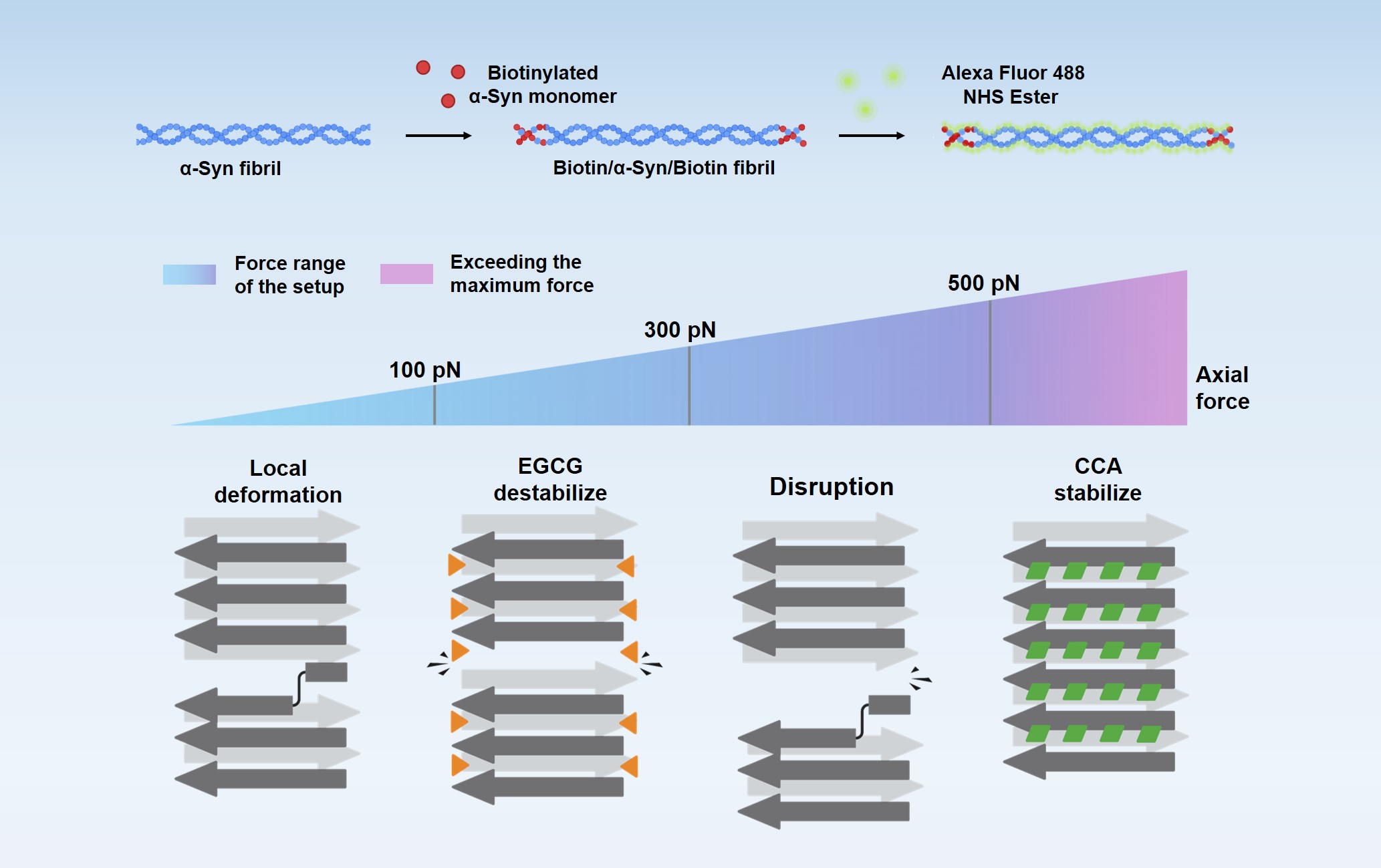 Physical characterization of polymorphic amyloid fibrils The aggregation of proteins into polymorphic amyloid fibrils and their deposition into plaques and intracellular inclusions are the hallmarks of amyloid diseases. We aim to uncover the mechanical dynamics of amyloid fibrils and their modulation by molecular chaperones and chemical compounds.
Physical characterization of polymorphic amyloid fibrils The aggregation of proteins into polymorphic amyloid fibrils and their deposition into plaques and intracellular inclusions are the hallmarks of amyloid diseases. We aim to uncover the mechanical dynamics of amyloid fibrils and their modulation by molecular chaperones and chemical compounds.
- per page 99 records total 5 records
- firstpage <<previouspage nextpage>> endpage
- PageNumber 1/1 jumpto